I study wildlife genetics, mostly. I bounce around between species and places.
CV: Williams_2025




Bringing the lab to the field, studying the interplay of genes and behavior in primates
Max Planck Institute of Evolutionary Anthropology, Leipzig Germany, October 2023 – present
I am currently based in Leipzig, Germany working in the Primate Behavior and Evolution group at the institute. We use genetic approaches to understand behavioral and life history events in primate populations. I’m learning various genetic and genomic protocols to later deploy in the field at our sites in Kenya, Uganda, and Costa Rica. We’ll be working with noninvasive samples collected from baboons, gorillas, and capuchin monkeys. Incredible advances in technology make it possible to sequence DNA from a backpack – using a small sequencing kit and device connected to a laptop! Deploying the genetic protocols in the field would provide faster results, less permitting to ship biological samples internationally, and provide the opportunity to train local researchers at the field sites.
This is a dream job for me where I can combine my nerdy science skills with my passion for travel and adventure. Challenges will involve troubleshooting genetic protocols that work well in sterile laboratories, but may be more difficult to carry out in a tent in Kenya or Uganda. In Costa Rica I will also be helping to renovate an abandoned building into a genetics lab (pic above), once I relocate the iguanas and bats currently inhabiting it. I will likely also be returning to South Africa to wrap up some of the Kalahari Meerkat and Molerat Project (see below for previous research). I’m very excited to bring the lab to the field and learn more about primates.




Environmental DNA to detect invasive species in a pristine sub-Antarctic archipelago
Cape Horn Biosphere, Chile, April 2023 – June 2023
The Cape Horn Biosphere Reserve, Chile, protects pristine sub-Antarctic ecosystems. However, this remote area lacks a long-term surveillance system of non-native species. Without an accurate account of invasive species distribution, it will be difficult to prioritize conservation efforts. Environmental DNA (eDNA) detection offers a sensitive, noninvasive method to detect and monitor species in the wild. Navarino Island has recently been invaded by the American mink and affected by growing populations of feral dogs; the island offers an ideal study system for developing an eDNA detection tool for invasive species in the Cape Horn Biosphere.
We spent three months trekking to remote water bodies (lakes and rivers) to deploy camera traps and collect DNA from water samples around the Cape Horn Biosphere. This work was part of an idea I’ve been working on since my time spent in the Kalahari. Due to the limited funding to cover field and lab supplies, I needed to be creative to make the study possible on a low budget. I designed a manual pump using a Nalgene water bottle, brake bleeding kit (pump and tubes), and self assembled funnels with 0.45 um filter paper. DNA will be extracted from the filters and a dPCR assay will be developed to detect DNA from mink and dog. The detection rates will be compared between eDNA sampling and camera traps. The work is ongoing.



Immune Cell Atlas of Indigenous South American Populations
South America, March 2022 – March 2023
I am currently traveling around South America as a field research technician for the Cell Atlas Project through the University of Chicago. We are interested in understanding the evolution of the immuno-genomic landscape of eight distinct indigenous populations from the Amazon, Andes, and coastal regions of South America. These populations encountered admixture from Eurasian and African sources, experienced founder events and genetic bottlenecks, and were exposed to selective pressures. We will compare shared and unique biological variation between populations in order to understand the immuno-genomic variation of these indigenous groups and the ways in which they are distinct to other ancestry groups.
I will be traveling between Chile, Peru, and Brazil to set up laboratory space to process blood samples collected from these populations. For each sample of 60 ml of blood, I will collect a genetic sample for the population genetics portion of the study. The majority of the blood will then be processed for isolating peripheral blood mononuclear cells (white bloods cells) for future antigen/virus challenges that will be carried out in Chicago to understand the immuno-genomics of these populations. Between each country sampling process, I fly with a liquid nitrogen tank full of the cell samples up to Chicago.



Associate in Research, studying the interplay of genes and behavior
Department of Evolutionary Anthropology, Duke University – Durham, NC Oct 2020 – July 2021
I focus on non-invasive genotyping and sequencing library preparation for paternity assignment and other genetic analyses of wild social mammals, particularly baboons. My duties include development, optimization, and execution of various wet lab protocols, including DNA extraction, high-throughput sequencing library preparation, reverse transcription, and DNA/library preparation control. I also provide project support and training to post-doctoral researchers, graduate students, and other lab technicians.
I ran my first MinIon sequencing run at Duke. This is the technology I’d like to use for my proposed study in Chile so it was really cool to be trained on it! The MinIon is a portable sequencing device that allows for real time long read sequencing of DNA. We are currently testing it in the lab to eventually be used at the Amboseli field station in Kenya to sequence baboon samples for questions about relatedness among individuals in the troops.


Human Frontier Science Program
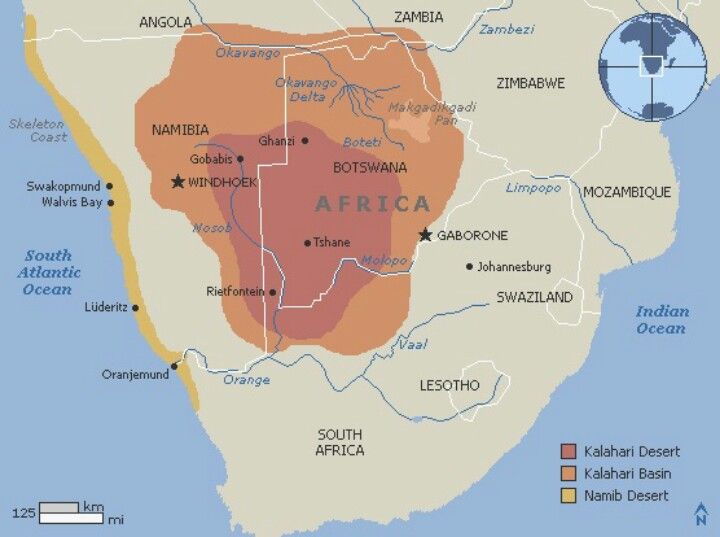
Kalahari Meerkat Project – Northern Cape, South Africa June 2019 – May 2020
I was the research manager of the cell laboratory affiliated with the Human Frontier Science Program at the Kalahari Meerkat Project. Research at the study site involves members of Duke University, the University of Chicago, and the University of Cambridge. We were looking at the effects of social dominance on immune responses of the South African Meerkat, Suricatta suricatta, and the Damaraland Molerat, Fukomys damarensis, by isolating peripheral blood mononuclear cells (PBMCs) from blood samples and adding various hormones and antigens as challenging reagents to assess immunological stress. Both species form cooperative societies where members of the group adopt alternative social roles to assist in the production of surviving offspring. For the meerkats, we hypothesized that competing demands such as reproduction, growth, and immune response, can be observed at the transcriptional (DNA) level and that these demands are resolved differently between levels of hierarchy within the population (helpers verses breeders). For the Damaraland mole rats, we studied how experimentally induced changes in dominance (queen) status influence the genomics of skeletal growth and aging. The “human frontier science” part comes in when we think about how human health is affected by social class. People of high social status tend to live longer than those in the lower classes. One reason may be wealthier individuals can afford better health care or working safer jobs. Studying gene regulation in cooperative animal societies allows an opportunity to ask this question at the genetic level – Does hierarchy affect DNA, and thus health?
The lab is located at a remote field station in the middle of the Kalahari Desert. Soon my duties will be expanded to include DNA extraction on site, and potentially DNA amplification (PCR) depending on the capacity of the space and resources we have here. I also set up a genetics lab next to the cell lab for future studies.
Publications:
A female-biased gene expression signature of dominance in cooperatively breeding meerkats

Meerkat Immune Responses
University of Chicago – May 2019 – June 2019
I trained in Chicago for the month to learn cell culture for my next position in South Africa. Basically, I isolated white blood cells from blood samples and challenge them with treatments to assess immune response. I practiced on my own blood first (which is pretty sweet) but will then I will be working with blood collected from meerkats. A group of meerkats is called a mob and there is one dominant breeding pair per mob. The rest of the mob helps raise the pups, this is called cooperative breeding. The project is interested in what distinguishes a dominant verses subordinate individual, both physically and genetically. Specifically what genetic changes are happening when a dominant breeder dies and a new subordinate takes his/her place. One way to understand this is by testing blood samples to see if immune responses change during turnover. I’ll also be working with molerats, less cute but pretty unique and amazing animals. When a molerat dies, I will be collecting various tissues and isolating bone marrow for stem cell research. I practiced on a mouse in Chicago as we lack molerats. Next stop – the Kalahari Desert!
Here is a link to the project: kalahari-meerkats.com/kmp/



Understanding Predator-Prey-Scavenger interactions through DNA – and various other projects
University of Washington, March 2017 – May 2019
I traded Colorado’s sun and brown for Washington’s rain and green. I built a wildlife genetics lab in a basement of one of the forestry buildings and helped graduate students complete the genetic components of their research projects. A common theme in many projects was determining carnivore species from clues left behind – like scat, saliva on bite wounds, hair, and snowtracks. I really liked this job because I could work on multiple projects and species, and I enjoyed teaching techniques in the lab. Other projects involved bee pollen, powdery mildew on plants, crow blood, nitrogen cycling bacteria, wood forensics, and pig gut microbe detection in water samples.
depts.washington.edu/sefsblog/tag/kelly-williams/
Publications:


Detection of environmental DNA from an invasive, terrestrial species – the feral pig
National Wildlife Research Center & Colorado State University, June 2013 – February 2017
My graduate research involved developing a test to detect the invasive feral pig by testing for DNA in water samples. Environmental DNA is shed from an animal into their environment via skin, hair, urine, feces, etc. The goal of this project was to develop a surveillance system on invasion fronts and in areas thought to be eliminated of feral pigs. Testing water sources (ponds/streams/water troughs) for shed DNA from feral pigs provides a noninvasive method for understanding species distribution. Once the method was optimized in the lab (dealing with challenges of turbid water samples), it was tested in the field in San Antonio, Texas for real world application.
Publications:
No filters, no fridges: a method for preservation of water samples for eDNA analysis
Clearing muddied waters: Capture of environmental DNA from turbid waters
Detection and persistence of environmental DNA from an invasive, terrestrial mammal

The start to it all – Undergrad research: Bone morphogenic protein and deer antler size
Niagara University, September 2009 – May 2013
My undergraduate research involved determining if various bone morphogenic proteins played a role in deer antler size. This research involved gathering tissue samples of deer from local taxidermists, isolating DNA from the tissue, and determining if the presence of certain genes played a role in antler size. My original plan was to go to vet school to become an equine veterinarian until I started working in Dr. Gallo’s lab and realized how much I like research and solving problems in the lab with creativity. I also learned how to pipette, that was probably also pretty important for my career path.